In response to your feedback, we’ve made more whole genome cross-species alignments available in NCBI’s Comparative Genome Viewer (CGV). You can use these alignments to explore genome rearrangements between species. You can also zoom in to analyze regions of conserved gene synteny.
There are over 20 new cross-species alignments available, including human-mouse, mouse-rat, human-chimp, human-cattle, dog-cat, and others! These cross-species alignments provide additional opportunities to explore evolutionary relationships at the genomic and gene levels. We will add more cross-species alignments in the coming months.
The latest cross-species alignments added to CGV include imports from the UCSC Genomics Institute, as well as those generated at NCBI.
Check out two examples of cross-species whole-genome alignments in CGV below (Figure 1).
Figure 1. Whole genome alignments between (A) mouse and human (GRCm39 vs. GRCh38.p14) and (B) cat and dog (F.catus_Fca126_mat1.0 vs. ROS_Cfam_1.0). Colored bands connects aligned regions; green indicates same orientation, blue indicates opposite orientation.
When you zoom in on an alignment (Figure 2), you can compare gene annotation on the two assemblies and see the extent of conservation of synteny. You can also see which genes are missing from one or the other assembly, indicating changes in sequence or differences in annotation.

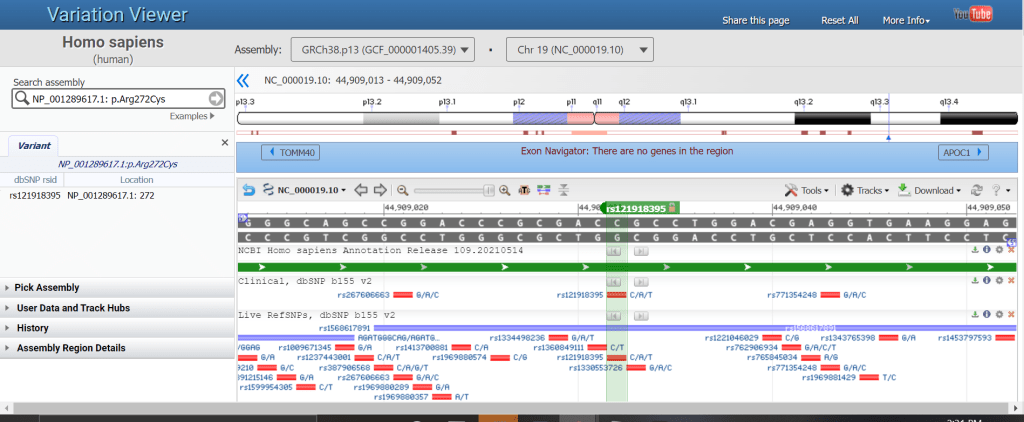


 Figure 1. The human ABO
Figure 1. The human ABO  Figure 1. A multiple alignment view in Genome Workbench highlighting the new ability to save presentation quality image files (Save As PDF and SVG formats).
Figure 1. A multiple alignment view in Genome Workbench highlighting the new ability to save presentation quality image files (Save As PDF and SVG formats). Figure 1. The Track Configuration panel showing the Remote Variation Data tab and he EVA RefSNP Release 1 track. Select the track checkbox and click Configure to load the track.
Figure 1. The Track Configuration panel showing the Remote Variation Data tab and he EVA RefSNP Release 1 track. Select the track checkbox and click Configure to load the track. Figure 2. The graphical sequence viewer showing the region of the growth hormone gene on pig chromosome 12 (
Figure 2. The graphical sequence viewer showing the region of the growth hormone gene on pig chromosome 12 (
