If you’ve ever tried searching for a genomic location in NCBI’s Genome Data Viewer (GDV) or Variation Viewer and found that your search term didn’t work, it’s time to try again! We recently expanded support for searches in our genome browsers using non-NCBI identifiers such as HGVS patterns (e.g. NM_001318787.2:c.2258G>A) and Ensembl IDs. You can also search by chromosome coordinates, cytogenetic band, assembly scaffold/component, disease/phenotype, dbSNP identifier, or RefSeq transcript/protein accession. We’ve gathered example searches in the table below.
| Search term | Example(s) |
|---|---|
| Chromosome coordinate | chr1:1,500,000-2,000,000 chr2: 1.5M-2,540.2K 3: 21.335M..21.337M 3: 21.335M..21.337M chr5 |
| Cytogenetic band | 1p36.21 2q13 |
| Assembly scaffold | NT_005403.18 NW_021159987.1 |
| Assembly component | AC106865.4 AC018680.4 |
| Gene/protein name | PTEN protease |
| Disease/phenotype | diabetes eye color |
| SNP rsID | rs863223352 |
| dbVar ID | rs863223352 |
| RefSeq transcript/protein accession | NM_017551.3 XP_011538173.1 |
| Ensembl gene/transcript indentifier | ENSG00000233258 ENST00000404547 |
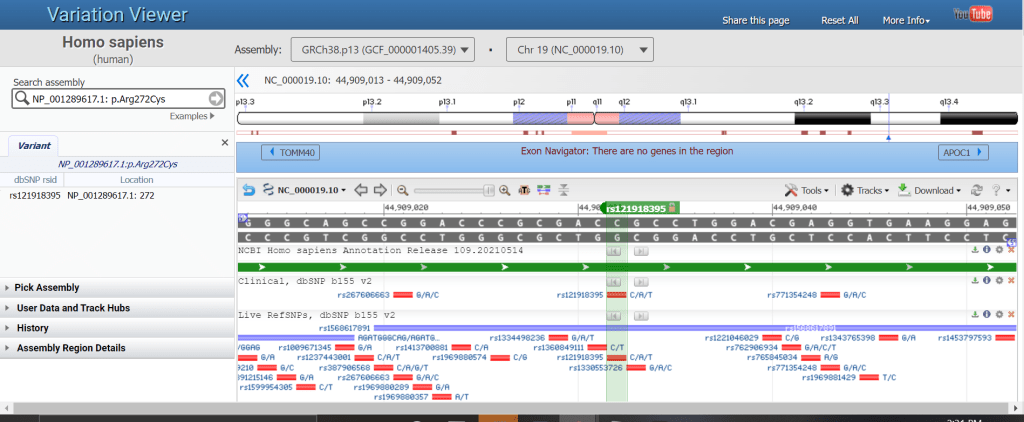
| HGVS | NM_001318787.2:c.2258G>A NP_001289617: p.Arg272Cys |
When you search by single coordinate, SNP or dbVar ID, or HGVS, the browser view zooms to the location of the search result. A marker is automatically created to identify the searched position. For HGVS, the marker is labelled with the corresponding rsID, if there is one.
As always, please contact us if you have additional questions or suggestions about this or any other feature in GDV or Variation Viewer. You can use the Feedback button on the page or write to the NCBI Help Desk directly.